Abstract
Background: Classification of interstitial lung disease (ILD) is challenging. Whole genome sequencing (WGS) results may be a novel biomarker aiding diagnosis.
Aim: To classify the lower airways microbiome in 11 patients with unspecific ILD compared with 12 idiopathic pulmonary fibrosis (IPF), 34 sarcoidosis and 100 healthy controls.
Methods: We sampled protected bronchoalveolar lavage from all participants. WGS was done with Illumina NovaSeq 6000, and bioinformatic analysis were performed using the Gaia pipeline.
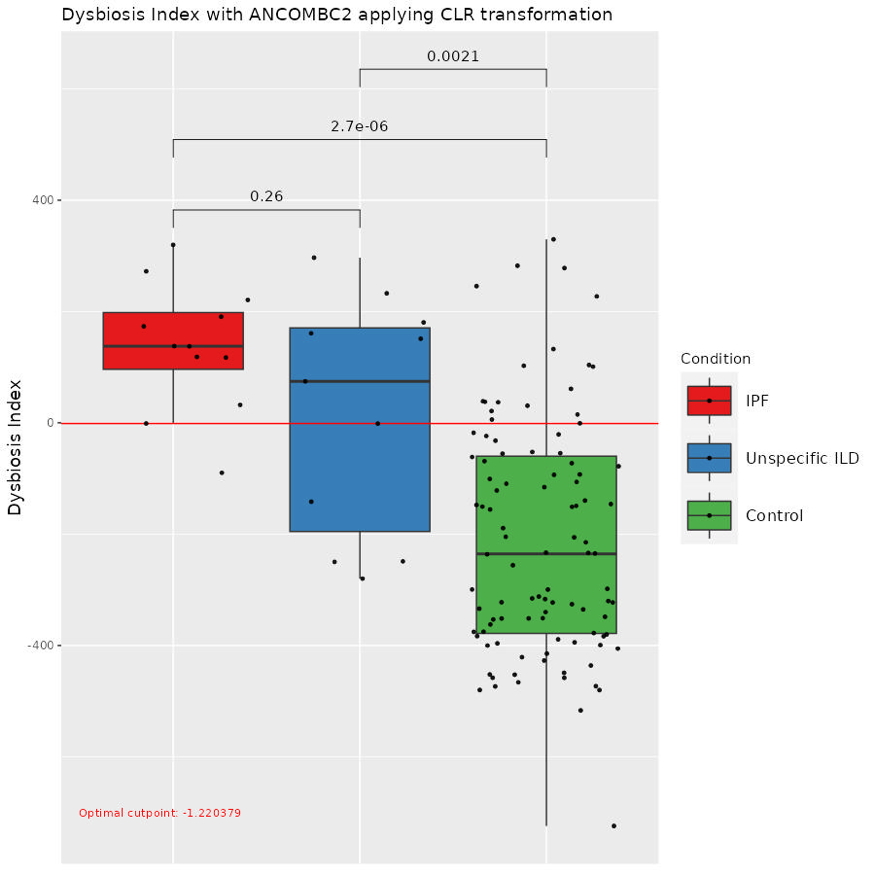
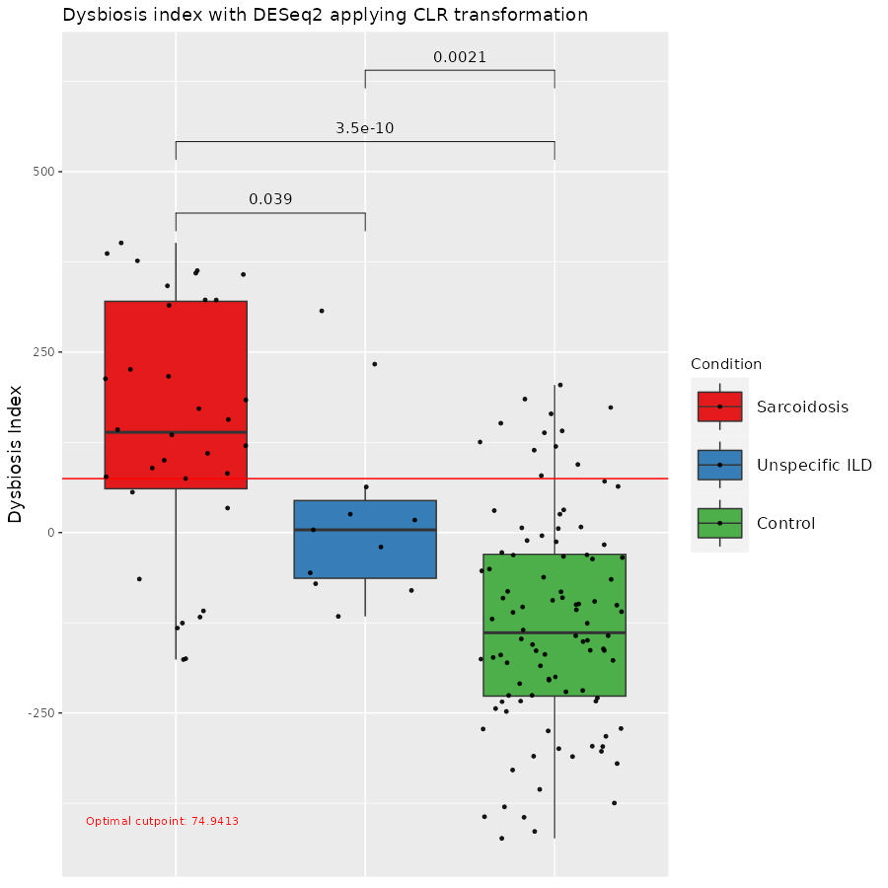
Results: Differential abundance analysis with ANCOMBC2 and DESeq2 identified several differentially abundant taxa. We calculated a dysbiosis index for each disease, comparing patients with healthy controls. The sensitivity and specificity for the index to differentiate between IPF and controls was 0.92 and 0.82, and respectively for sarcoidosis and controls 0.79 and 0.86. Applying the indexes on patients with unspecific ILD, we could discriminate against sarcoidosis for 10 patients, and indicate a possible similarity with IPF in 6 patients (Fig 1&2).
Conclusion: Lung dysbiosis index may be a novel biomarker in diagnosis of interstitial lung disease.