Abstract
Introduction
Pulmonary manifestations of COVID-19 range from asymptomatic cases over severe acute lung injuries such as ARDS to pulmonary long-COVID manifestations. In addition to hyperinflammation, fibrotic lung changes may occur, whose pathogenesis is not yet fully understood.
Aims and objectives
The aim of our pilot study was to characterize differences of fibrosis-associated pathways in patients with acute COVID-19 ARDS and long-COVID-ILD compared to non COVID-19 ARDS and other ILD, respectively.
Methods
We performed comprehensive transcriptome analyses for RNA sequencing (TruSeq-RNA Kit, Fa. Illumina) on 18 formalin-fixed, paraffin-embedded (FFPE) lung samples (autopsy/intraoperative tissue samples, lung biopsies) from patients with COVID-19 ARDS +/- invasive ventilation and long-COVID-ILD compared to 30 control samples from non COVID-ARDS, NSIP, COP, IPF and healthy lung tissue.
Results
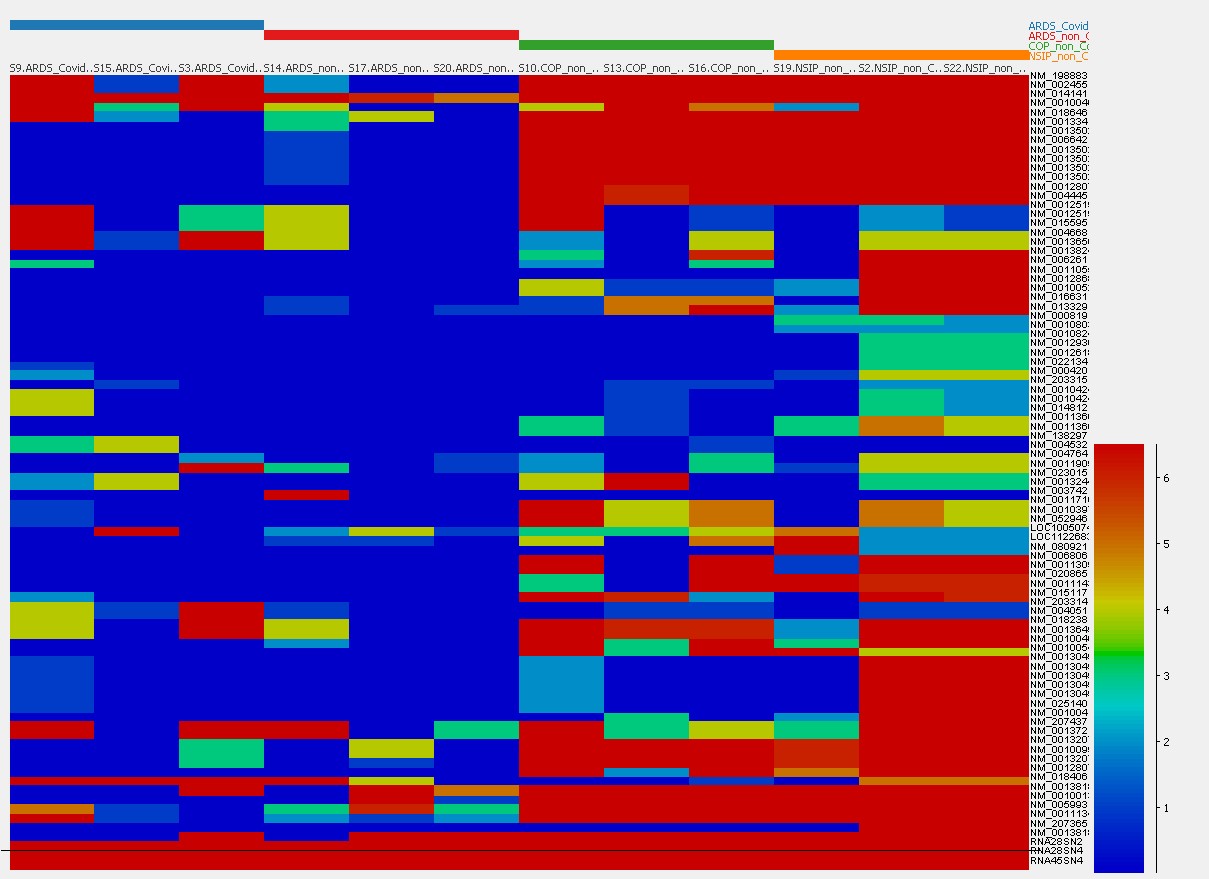
In general, transcriptome analyses on FFPE material were challenging due to RNA denaturation. Both homologous and significantly differentially regulated gene expression patterns were identifiable relating to specific fibrosis pathways in COVID-19, non COVID-ARDS, COP and NSIP (see Figure 1).
Conclusions
Our preliminary transcriptome analyses suggest distinct gene expression patterns in the various subgroups and may set the basis for future molecular biological studies to understand specific lung fibrosis pathways.