Abstract
Introduction: Idiopathic pulmonary fibrosis (IPF) and fibrotic hypersensitivity pneumonitis (fHP) are among the most common etiologies underlying pulmonary fibrosis. Data regarding molecular differences underlying these two entities is lacking.
Objectives: To assess the differentially expressed genes (DEGs) and pathways between fHP and IPF broncholaveolar lavage (BAL) cells by performing single-cell RNA-sequencing (scRNA-seq).
Methods: ScRNA-seq was performed on BAL samples from 2 patients with IPF, 2 fHP, and 1 healthy control. Computational analysis was performed using Cell Ranger and Seurat. DEGs and pathway analysis were performed on this data, as well as on integrated previous lung scRNA-seq datasets from fHP and IPF patients for comparison.
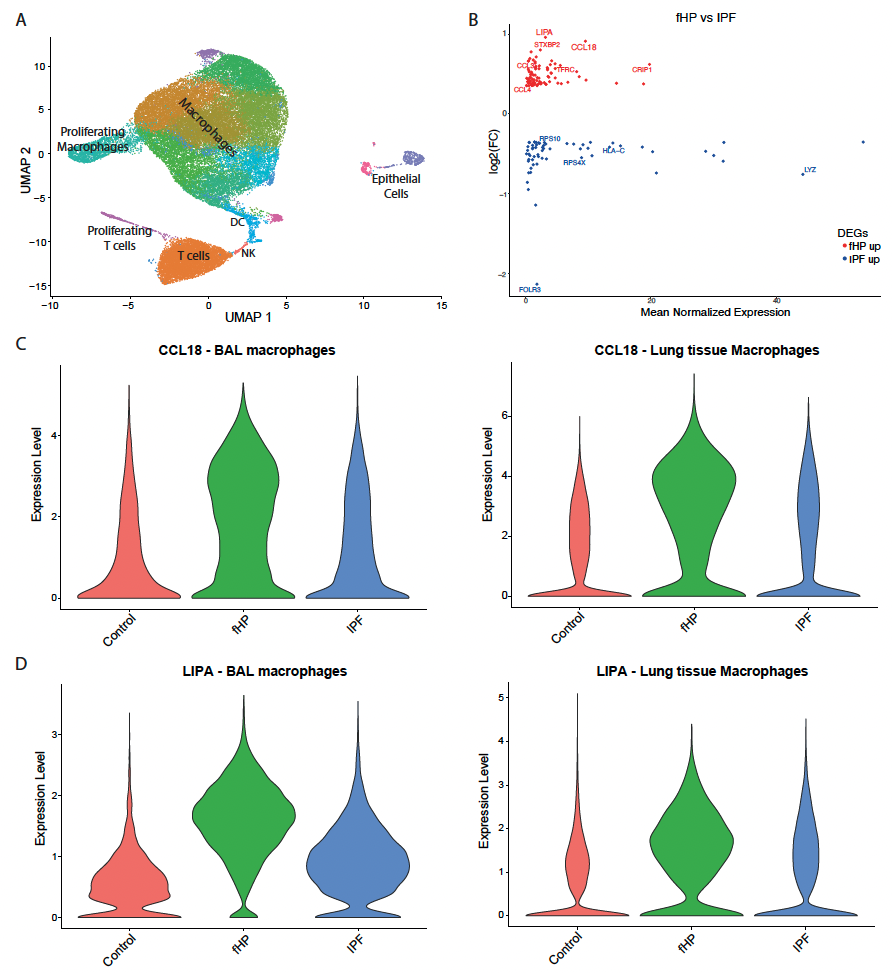
Results: A total of 27072 cells were subjected to unsupervised clustering, yielding 8 macrophage clusters (Fig. 1A). Compared to IPF, macrophages of fHP patients demonstrated a pro-inflammatory activation pattern including increase in CCL2, CCL3, CCL4, CCL18, and CXCL8 (Fig. 1B). A similar pro-inflammatory signature was detected in the lung scRNA-seq datasets. LIPA, a lysosomal acid lipase, was increased in fHP vs IPF macrophages in both datasets (Fig. 1C-D).
Conclusions: A pro-inflammatory signature was detected in fHP macrophages compared to IPF, which may translate to a diagnostic biomarker to help differentiate these entities.