Abstract
INTRODUCTION: COVID19 bimodal clinical and pathological phenotypes was characterized by fibrotic and thrombotic poles as previously described by our group.
AIMS AND OBJECTIVES: To correlate the COVID19 bimodal phenotypes into their transcriptomic profile.
METHODS: A retrospective study was carried out with 13 frozen lung tissue samples collected in minimally invasive autopsies, RNA extraction and NGS(Thermo) at the HiSeq Illumina. Bioinformatics was analyzed with RNASeq pipeline, heatmap and differential expression.
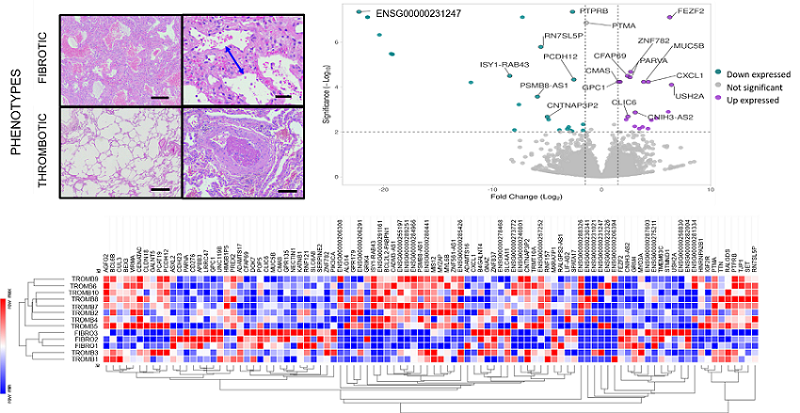
RESULTS: 606 genes were differentially expressed between the both phenotypes (P<0.05)(Fig1). The fibrotic phenotype up-expressed genes were related to extracellular matrix proteins as PARVA, MUC5B and USH2A. On the other hand, genes as PCDH12, PTPRB, and a non-identified gene may be related to thrombotic and vascular injuries, as highlighted by up-expressed genes in thrombotic phenotype. Clustering the 100 most differentially expressed genes resulted in 2 phenotypes(Fig1).
CONCLUSIONS: The whole transcriptome revealed genes related to extracellular matrix in fibrotic phenotype and endothelial cells proliferation in thrombotic phenotype, according to our previous bimodal phenotypes. Correlating these genes with patients? outcomes and survival are the following steps to elucidate their molecular pathways and biomarkers.
FINANCIAL SUPPORT: Research Support Foundation of São Paulo State (FAPESP Nº21/09024-6,20/13370-4, 22/02821-0).