Abstract
Introduction: COPD patients with high levels of blood eosinophils have a better therapy response to corticosteroids, but also an increased risk of exacerbations. We aimed to evaluate the lung bacterial microbiome related to eosinophil levels.
Methods: Bronchoalveolar lavage was sampled from the right middle lobe of 91 COPD patients with bronchoscopy. Blood-eosinophils were measured, cut-off between high vs low levels was set at 0.3 * 10^9 cells/L. Whole genome sequencing was performed on the Illumina NovaSeq platform. Identification of taxa was made by the GAIA 2.0-software. Statistical analyses were performed in R (Phyloseq-, DeSeq2- and AncomBC-package).
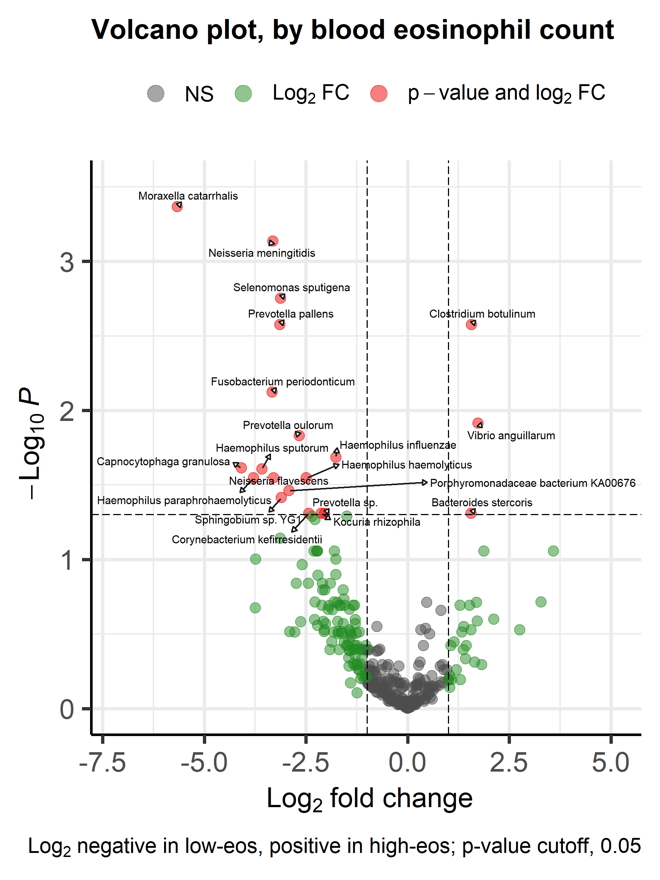
Results: Alpha diversity (Shannon and Simpson) and Beta diversity (Bray-Curtis dissimilarity) were not significantly different between the groups. With DeSeq2, 21 species were differentially abundant; 17 species in low-eos COPD, top 4 were Moraxella catarrhalis, Capnocytophaga granulosa, Haemophilus paraphrohaemolyticus and Haemophilus sputorum, whereas 4 species were more prevalent in high-eos COPD; Clostridium tetani, Vibrio anguillarum , Clostridium botulinum and Bacteroides stercoris. With AncomBC, only Moraxella catharrhalis was differentially abundant and more prevalent in low-eos COPD.
Conclusion: Overall, the lung microbiome is similar in low- and high-eos COPD, but several clinically significant species are differentially abundant.